Multiple Choice
Identify the choice that best completes the
statement or answers the question.
|
|
|
The following diagram represents the Lac operon in two different
environments. Answer the following questions based on this diagram.
|
|
|
1.
|
RNA polymerase must bind:
a. | 3 | b. | 4 | c. | 5 | d. | 7 | e. | Both (4) and
(7) |
|
|
|
2.
|
The advantage of the Lac operon is:
a. | The expression of genes can be regulated by environmental
signals. | b. | Proteins not necessary in a specific environment will not be
expressed. | c. | Human cells do not have to make all proteins encoded by its DNA. | d. | Both (a) and
(b). | e. | All of the above. |
|
|
|
3.
|
The repressor protein can bind:
a. | 3 and 2 | b. | 5 and 6 | c. | 4 and
9 | d. | 5 and 9 | e. | 6 and 9 |
|
|
|
4.
|
The molecule labeled (8) is:
a. | Lactose | b. | Ribosome | c. | Helicase | d. | DNA polymerase | e. | RNA
polymerase |
|
|
|
5.
|
The molecules labeled (10) are used by the bacteria to:
a. | Break down lactose to generate ATP through cellular respiration. | b. | Bind the operator to
inhibit transcription. | c. | Activate RNA polymerase by breaking down the
repressor molecule. | d. | Begin DNA replication. | e. | Create additional
sugars for the bacteria. |
|
|
|
The following plasmid map shows the three genes on the plasmid pBIO. The
GFP gene and ampicillin resistance gene ( AMPR) are regulated by a promoter
that is always “on”. Therefore, the genes are always expressed once in a
cell. The kanamycin resistance gene ( KANR) is regulated by the Lac
operon. Kanamycin is another antibiotic that prevents the grow of bacteria. Plates
labeled with “Amp” and “Kan” contain the corresponding antibiotics. The
plate labeled with “lactose” contains the sugar lactose. A bacterial culture is
transformed with pBIO and successfully transformed cells are plated on all four plates shown
below. Answer the questions based on your understanding of transformation, regulation by the
Lac operon, and gene expression.
|
|
|
6.
|
You transform bacteria (E.coli) with the plasmid pBIO. The successfully
transformed cells are plated onto the four plates above. On which plate will you not see
any growth?
a. | LB | b. | LB/Amp | c. | LB/Amp/Kan | d. | LB/Amp/Kan/lactose | e. | All plates will have
some growth |
|
|
|
7.
|
Which of the following is not an observation you will make?
a. | The LB plate will show lawn growth because all plated bacteria, including those that
did not take up a plasmid, will be able to grow. | b. | The LB/Amp plate will show colony growth.
Each colony represents a single bacterium that took up the plasmid pBIO and grew into millions of
cells during incubation. | c. | All colonies on the LB/Amp/Kan/Lactose plate
will GLOW because GFP is expressed. | d. | The colonies on the LB/Amp plate will
GLOW. | e. | When untransformed E.coli is plated onto LB/Amp/Kan/lactose, you will see lawn growth
because lactose will help make bacteria resistant to the antibiotics. |
|
|
|
8.
|
Which of the following is false about the plasmid pBIO?
a. | It is a circular piece of DNA. | b. | It codes for enzymes that make bacteria
resistant to antibiotics. | c. | It codes for enzymes that make the antibiotic
kanamycin. | d. | Contains three genes. | e. | It codes for a gene that makes a protein that
will fluoresce. |
|
|
|
9.
|
The process illustrated in the above diagram is ----
a. | transformation- bacterial cell are induced to uptake a new plasmid. | d. | asexual
reproduction- bacterial cells are replicating and dividing | b. | transillumination-
bacterial cells are made competent. | e. | sexual reproduction- bacterial cells are exchanging genes. | c. | horizontal transfer
of genetic material between bacterial cells. |
|
|
|
10.
|
Referring to the diagram which answer best illustrates the explanation of the
process between step 1 and step 3?
a. | The addition of a selectable marker to media | d. | The uptake of the plasmid into
all bacterial cells | b. | The addition of a selection pressure on the
bacterial phenotype | e. | The
addition of LB for the recovery phase of transformation | c. | The uptake of the
plasmid into a bacterial cell |
|
|
|
Below is the plasmid map of pAMP. The plasmid has a single ampicillin
resistance gene and an origin of replication. Restriction sites and corresponding nucleotide
numbers are shown. The entire size of pAMP is 4539 base pairs (bp).
The plasmid was cut with the enzymes BamHI and EcoRI. This
reaction produced two fragments with sizes 1120bp and 3419bp.
You accidentally mix up the
digestion reaction with another that you were using for a separate experiment (this is why you should
always label tubes!). Unable to figure out which tube contained your original digestion
reaction, you run samples of both tubes in a gel along with a ladder.
The ladder standards are
DNA fragments with the following sizes: 5000, 4000, 3000, 2000, and 1000.
The diagram to the
right shows the resulting gel. |
|
| |
|
|
|
11.
|
Which of the following statements are true?
a. | Lane 1 contains pAMP digested with BamHI and EcoRI. | b. | Lane 2 contains pAMP
digested with BamHI and EcoRI. | c. | This gel does not provide sufficient
information to identify your pAMP digestion. | d. | Without knowing how much DNA was loaded into
each well in the gel, it is not possible to determine which lane contains your pAMP
digestion. | e. | Both lane 1 and 2 seems to contain the same sample. |
|
|
|
12.
|
You prepare another restriction digestion and digest pAMP with the enzyme
BglI. When you run this DNA sample in a gel, how many bands should you see?
|
|
|
The following diagram describes how the
human insulin gene can be cloned into a plasmid and transformed into bacteria to produce human
insulin for therapeutic use. Steps 1 through 3 describe essential steps necessary to create a
copy of the gene that can be expressed in bacteria. Each numbered step is a enzyme-facilitated
step. Answer the following questions based you your understanding of the
diagram.
|
|
|
13.
|
Why are steps 1 through 3 necessary to create a version of the insulin gene that
can be expressed in bacteria? In other words, why can’t the original gene simply be cut
out from human DNA and inserted into a plasmid?
a. | Bacteria do not have mechanisms to splice introns. | b. | The gene must be
transcribed into RNA, which can then be inserted into the plasmid. | c. | The original gene
lacks the 5’ cap and 3’ poly-A tail necessary for gene expression in
bacteria. | d. | Both (a) and (b) above. | e. | All of the
above. |
|
|
|
14.
|
Step 2 represents:
a. | Transcription. | b. | Removal of introns. | c. | Translation. | d. | Replication. | e. | Digestion by
restriction enzymes. |
|
|
|
15.
|
The enzyme used in step 3:
a. | is reverse transcriptase. | b. | is RNA polymerase. | c. | uses RNA as a
template to create double stranded DNA. | d. | Both (a) and (c). | e. | Both (b) and
(c). |
|
|
|
16.
|
The enzymes used in step 4 is (are):
a. | DNA polymerase and ligase. | b. | EcoRI and HindIII. | c. | Ampicillin and
ORI. | d. | RNA polymerase. | e. | Reverse
transcriptase. |
|
|
|
17.
|
After bacteria is transformed with the recombinant plasmid (step 6), the culture
was plated on the two plates shown in the diagram (LB/Amp and LB/Kan). Which of the following
statements is true?
a. | There will be colony growth on both plates. | b. | The bacteria growing
on the LB/Kan plate contains the recombinant plasmid and is capable of producing human
insulin. | c. | Only the LB/Amp plate will have colony growth. | d. | Only the LB/Kan
plate will have colony growth. | e. | There will be lawn growth on the LB/Amp
plate. |
|
|
|
The following questions refer the series of diagrams below. Numbers 1, 3, and 5
refer to the structures in the diagram. Number 2, 4, and 6 refer to step in the process of
restriction digest of a plasmid
|
|
|
18.
|
The process between step 1 and step 3 is best explained by which of the
following statements.
a. | The DNA of two different strains of bacteria is being ligated
together | b. | The addition of a restriction enzyme to the reaction has created complementary sticky
ends on the plasmid and insert | c. | The addition of an antibiotic has cut the
plasmid in two random sites | d. | The DNA has been heated to 97o to unzip the
double stranded DNA | e. | The DNA backbone has been nicked and primers
will anneal to the open region |
|
|
|
19.
|
Refer to the difference between structure above number 5 and the structures
above number 7. For this amplification process to occur which of the following choices is
(are) corrent?
a. | The plasmid in step five would need to be digested with several restriction enzymes
and then ligated on to the surface recognition proteins of the bacterial cell for
replication. | b. | The plasmid above number 5 would require the process of transformation into a host
bacterial cell. The cellular machinery of the cell would then replicate the new
plasmid. | c. | The plasmid would need to be attentuanted with a dose of
antibiotics | d. | PCR the plasmid with taq polymerase at 73o. | e. | PCR with primirs,
taq polymerase, and free nucleotides. |
|
|
|
20.
|
The step between 3 and 4 on the diagram shows an opened plasmid. Which statement
best describes how the plasmid was opened?
a. | restriction digest with restriction enzymes | b. | PCR- polymerase
chain reaction | c. | mini prep- to remove the plasmid | d. | reverse transcriptase | e. | none of the
above |
|
|
|
|
|
|
21.
|
The above image shows a plasmid digest with Ava I enzyme on three different
plasmids.
Which of the following explanation best describes the results observed from the
gel.
a. | Lane 1 shows a plasmid with possibly one cut site | b. | Lane 1 shows a
plasmid with two possible cut sites | c. | Lane 2 shows a digest with two cut
sites | d. | Lane 3 shows a plasmid with three cut sites | e. | All of the above are
correct |
|
|
|
2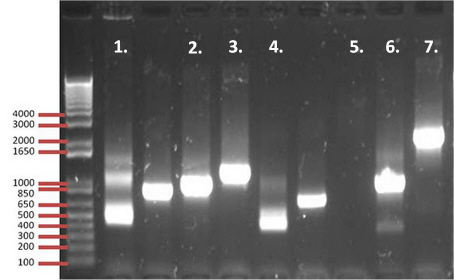
|
|
|
22.
|
Which lane has the biggest fragment?
|