Pictured: B. Carlson, S. Contess, A. Feng, V. Oliveira, V. Wei, A. Young, K. Kolb, S. Hanchuk with Ann Stock, Ph.D. UMDNJ, Piscataway, HHMI
To see previous Pingry SMART Team projects please click here
Pingry S. M. A. R. T Team 2008-2009
Students Modeling a Research Topic
coordinated by the Center for BioMolecular Modeling, Milwaukee School of Engineering, Milwaukee, WI
Tim Herman, Ph.D, Director
Teacher Advisors- Deirdre O'Mara, Tommie Hata
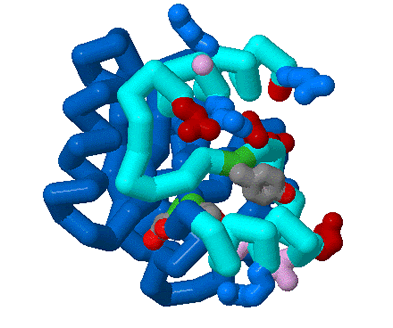
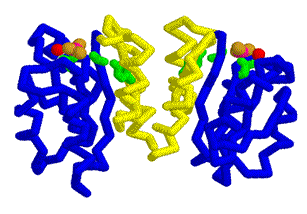
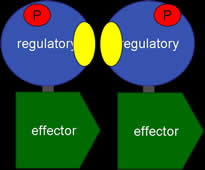
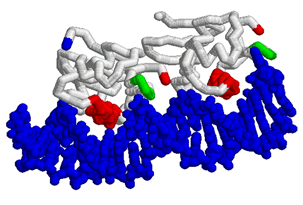
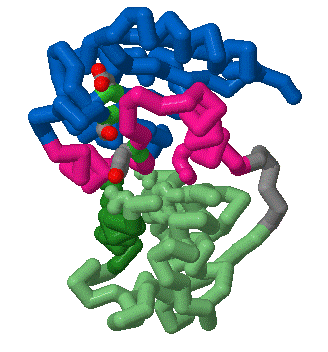
This year we are working with Anne Stock (HHMI, Center for Advanced Biotechnology, UMDNJ, Rutgers University, Piscataway, NJ) to create three dimensional models using RP-Rasmol of several proteins involved in the signal transduction pathway of bacteria. The proteins we intend to model are the second component of a two component signal transduction mechanism common in prokaryotes. The first half of the signal transduction pathway results in the phosphorylation of a protein kinase which activates a response regulator. We intend to model this response regulator protein from various species in different states of activation. The response regulator protein consists of two domains- the regulator region and an effector region. The regulator component, when phosphorylated, will dimerize in a symetrical formation. The regulator domain, connected via flexible linker to the effector domain will bind to DNA in a winged helix motif and in a directional orientation.
The SMART Team experience begins with the selection of a mentor for our students and reading the primary literature related to the proteins of interest. With guidance from our mentor, reading the literature students create virtual three dimensional models of the proteins using RasMol. These files then are engineered for three dimensional printing on a Z-Corp machine at MSOE.
Th culmination of the project is a poster presenation at the international meeting of American Society for Biochemistry and Molecular Biology (ASBMB) held this year in New Orleans, LA